Dear readers,
Today, I took some data on the length of the longest axis (mm) of the leaves and the length of the complementary axis (mm) of the leaves, so that I can measure (by a ruler) physical growth of the leaves.
Besides physical growth, I also measured the weight of two largest leaves (in grams) by harvesting (with scissors) two leaves from each plant.
For fitting the regression line, I took the average of the weights of two leaves. I wanted to know whether there is correlation between the weight of the two largest leaves and length of the axis.
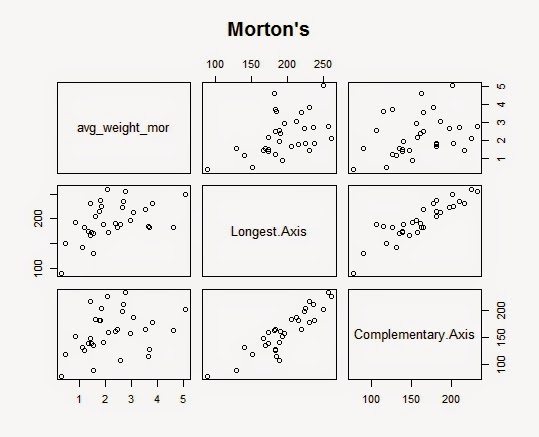
In Morton's Mix, we see a very strong correlation between the weight and the longest axis and also between the two axises. However, a relationship between the weight and the complementary axis looks mild. So, let's check the summary table for regression line of Morton's Mix.
> summary(mmod)
Call:
glm(formula = avg_weight_mor ~ Longest.Axis + Complementary.Axis,
data = mor)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.4458 -0.5931 -0.1255 0.6400 2.8105
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.92087 0.95130 -0.968 0.3410
Longest.Axis 0.02926 0.01047 2.795 0.0091 **
Complementary.Axis -0.01586 0.01023 -1.551 0.1317
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for gaussian family taken to be 0.9603569)
Null deviance: 39.636 on 31 degrees of freedom
Residual deviance: 27.850 on 29 degrees of freedom
AIC: 94.368
Number of Fisher Scoring iterations: 2
From this we can see that complementary axis does not fit well in this regression line, since the p-value for the hypothesis test (null: Beta3=0, alternative: Beta3 does not equal 0) is greater than the alpha level (0.05).
However, as I mentioned above, it might be possible to see a correlation between the two axises. So, let's try to fit a regression line for them.
> summary(mmod.2)
Call:
glm(formula = Longest.Axis ~ Complementary.Axis, data = mor)
Deviance Residuals:
Min 1Q Median 3Q Max
-33.709 -8.752 -3.709 12.502 39.974
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 55.61310 13.12115 4.238 0.000198 ***
Complementary.Axis 0.87302 0.07999 10.914 5.74e-12 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for gaussian family taken to be 292.1042)
Null deviance: 43557.0 on 31 degrees of freedom
Residual deviance: 8763.1 on 30 degrees of freedom
AIC: 276.41
Number of Fisher Scoring iterations: 2
Here, the result is as we expected. So, instead of treating the two axises as separate variables, let's combine the two and take a mean of it.
> summary(mmod2)
Call:
glm(formula = avg_weight_mor ~ avg_axis_mor, data = mor)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.4144 -0.7500 -0.2386 0.5838 2.4391
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.024277 0.919078 -0.026 0.9791
avg_axis_mor 0.012842 0.005078 2.529 0.0169 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for gaussian family taken to be 1.089074)
Null deviance: 39.636 on 31 degrees of freedom
Residual deviance: 32.672 on 30 degrees of freedom
AIC: 97.477
Number of Fisher Scoring iterations: 2
Here, we can clearly see that the weight of the leaves and the axises are correlated and now the variable for axises fits the data well.
Now, let's look at Freedom's Mix.
Here, it looks like all three variables are correlated.
> summary(fmod)
Call:
glm(formula = avg_weight_free ~ Longest.Axis + Complementary.Axis,
data = free)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.8606 -0.4031 -0.1565 0.3417 1.3652
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -1.328302 0.683507 -1.943 0.06173 .
Longest.Axis 0.015163 0.004990 3.039 0.00499 **
Complementary.Axis 0.002875 0.003717 0.773 0.44551
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for gaussian family taken to be 0.3716524)
Null deviance: 21.566 on 31 degrees of freedom
Residual deviance: 10.778 on 29 degrees of freedom
AIC: 63.989
Number of Fisher Scoring iterations: 2
However, we can see that the complementary axis does not fit the data well according to the p-value shown above.
So, let's combine the two axises again and take the mean of it.
> summary(fmod2)
Call:
glm(formula = avg_weight_free ~ avg_axis_free, data = free)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.8537 -0.4079 -0.1950 0.2780 1.2990
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.715077 0.561917 -1.273 0.213
avg_axis_free 0.015976 0.003155 5.064 1.95e-05 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for gaussian family taken to be 0.3875975)
Null deviance: 21.566 on 31 degrees of freedom
Residual deviance: 11.628 on 30 degrees of freedom
AIC: 64.418
Number of Fisher Scoring iterations: 2
As we expected, now the axises fit the data well.
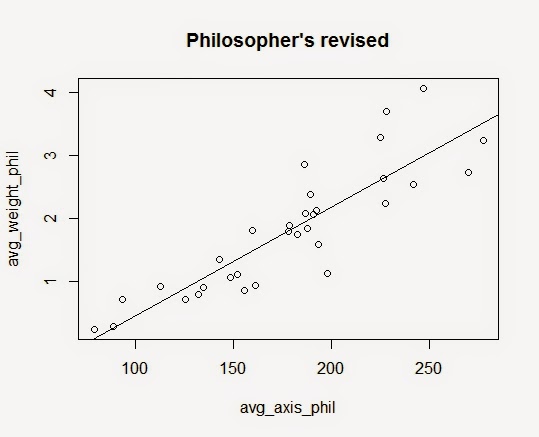
Now, let's look at the plot for Philosopher's Mix.
Here again, it seems like all three variables are strongly correlated to each other.
> summary(pmod)
Call:
glm(formula = avg_weight_phil ~ Longest.Axis + Complementary.Axis,
data = phil)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.99924 -0.29086 -0.00653 0.18672 1.07923
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -1.611472 0.294235 -5.477 6.75e-06 ***
Longest.Axis 0.017902 0.002865 6.248 8.08e-07 ***
Complementary.Axis -0.001399 0.003081 -0.454 0.653
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for gaussian family taken to be 0.17441)
Null deviance: 30.1881 on 31 degrees of freedom
Residual deviance: 5.0579 on 29 degrees of freedom
AIC: 39.779
Number of Fisher Scoring iterations: 2
However, we again see that the complementary axis does not fit the data well. But, we know (I checked) that the two axises are correlated.
So, let's combine the two axises and take the mean of it.
> summary(pmod2)
Call:
glm(formula = avg_weight_phil ~ avg_axis_phil, data = phil)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.0221 -0.2485 -0.0054 0.1687 1.0776
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -1.273861 0.320245 -3.978 0.000406 ***
avg_axis_phil 0.017252 0.001734 9.951 5.14e-11 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for gaussian family taken to be 0.2339939)
Null deviance: 30.1881 on 31 degrees of freedom
Residual deviance: 7.0198 on 30 degrees of freedom
AIC: 48.268
Number of Fisher Scoring iterations: 2
Now, we see that the axises fit the data well.
p.s. the root growth looks pretty good too.